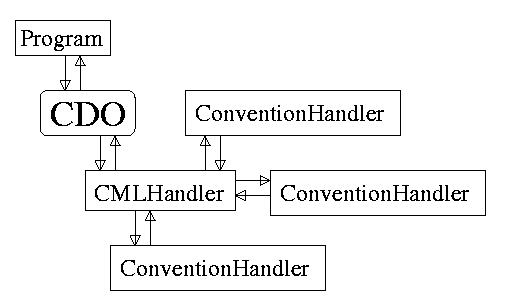
Figure 1: the CML import filter consists of a main handler that dynamically uses convention handlers to parse a CML file. Programs call upon this CMLHandler and use a chemical data object (CDO) as an interface.
CML is an XML language and was developed by P.Murray-Rust and H.S.Rzepa to store chemical information 2. In May 1999 they release the first specification of this versatile markup language. Since then two chemical programs started to support CML: JMol 3 and JChemPaint 4. Jumbo was the first the first program that is able to handle CML documents and also PDB to CML conversion tool available 2a, while a more comprehensive conversion tool is underway.
For both JMol and JChemPaint import and export were written earlier this year, but last month these filters turned out to be inconvenient. The reason these filters were not sufficient was that CML is extremely adaptable, i.e. the document type definition does not contain a convention on element dependencies nor on the meaning of data: "In this article we very deliberately do not attempt to develop or reconcile chemical ontologies."
For example, the bond order could be represented by the numbers one, two, three. Other types of bonds could be represented by four (aromatic bond) and five (hydrogen bond). But without any notice of a convention one could not determine what four and five could mean. In CML the default convention can be overwritten with the attribute mentioned above.
Consider the next CML file which was converted from PDB with the PDB2CML conversion tool 2a.
<?xml version="1.0"?>
<!DOCTYPE list SYSTEM "cml.dtd" >
<!--CML conversion of file 75-09-2.pdb
Produced by PDB2CML (c) Steve Zara 1999-->
<list title="molecules" convention="PDB">
<list title="model">
<list title="compounds">
<string title="compound"></string>
</list>
<list title="sequence" id="1">
<list title="" id="1">
<string builtin="residueType"></string>
<atom title="atom" id="1">
<string title="name">C</string>
<coordinate3 builtin="xyz3">-0.254 -0.572 0.101</coordinate3>
<float builtin="occupancy">1.00</float>
<float title="tempFactor">0.00</float>
</atom>
<atom title="atom" id="2">
... etc ...
</atom>
</list>
<feature title="ter" id="6"/>
</list>
<list title="connections"/>
</list>
</list>
To preserve all the data that was contained in the PDB will S.Zara developed this CML PDB convention. Not only does it use specific features of PDB, like PDB's TER command, but the whole structure of this CML file is specific for the PDB convention. As can be seen the file has a specific structure of molecules-model- sequence-atoms. In the CML file this structure is maintained with the use of the CML's list elements.
A similar thing is done in the JMOL-ANIMATION convention. Each frame of the animation consists of a molecule structure which are all a child element of a list element:
<?xml version="1.0" encoding="ISO-8859-1"?>
<!DOCTYPE molecule SYSTEM "cml.dtd" [
<!ATTLIST list convention CDATA #IMPLIED>
]>
<list convention="JMOL-ANIMATION">
<molecule id="FRAME1">
<string title="COMMENT">HEAT OF FORMATION = 38.45792 KCAL = 160.90796 KJ; FOR REACTION COORDINATE = 3.00000 ANGSTROMS</string>
<atomArray>
<stringArray builtin="id">
a0 a1 a2 a3 a4 a5 a6 a7 a8 a9 a10 a11 a12 a13 a14 a15 a16 a17 a18 a19 a20 a21 a22 a23 a24
</stringArray>
<stringArray builtin="elementType">
C C C C C H C H H H H H H O H H O H H H H H H H H
</stringArray>
<floatArray builtin="x3">
... a lot of data ...
</floatArray>
<floatArray builtin="y3">
... a lot of data ...
</floatArray>
<floatArray builtin="z3">
... a lot of data ...
</floatArray>
</atomArray)
</molecule>
<molecule id="FRAME2">
</molecule>
... etc ...
</list>
With these convention the processing unit is able to determine wether or not these fragements are seperate frames of an animation or if they are fragments of a complex. But now an old problem resurfaces. In the near future a lot of conventions will appear. Old ontologies will use new conventions, and new programs will use new conventions to be able to store chemical information they are not able to do or do not want to do in other conventions. Therefore, S.Zara and I proposed a documention system to store information on these conventions like element dependencies and more 5.
SAX compliant parsers use an event-based API, i.e. when the start of an element is encountered it signals a handler that processes the document with an startElement event. Other events are endElement, CharacterData and start- and endDocument. More information can be found at the Sax website 8.

The JMol and JChemPaint import filters consists of an interface with the chemical program (the CDO), a CMLHandler that dynamically uses an appropriate convention handler to process the CML file and store the data in the CDO. The CDO is an object that is specific for one program. Both JMol and JChemPaint have a seperate CDO. The model can even extend from an object that is already provided by the program. The only restriction is that the object should implement the CDO programming interface 9.
Consider the next Java code taken from JMol:
public CMLFile(InputStream is) throws Exception {
super();
String pClass = "com.microstar.xml.SAXDriver";
InputSource input = new InputSource(is);
Parser parser = ParserFactory.makeParser(pClass);
EntityResolver resolver = new DTDResolver();
DocumentHandler handler = new CMLHandler((CDOInterface)new JMolCDO());
parser.setEntityResolver(resolver);
parser.setDocumentHandler(handler);
parser.parse(input);
frames = (JMolCDO)((CMLHandler)handler).returnCDO();
System.out.println("Back in CMLFile...");
}
A program that want to use the CML import filter should define a XML parser as is done in line 6 and if the document is to be checked for validity a DTDResolver. The handler that the parser uses actually processes the XML document and is of course the CMLHandler in our case. The CMLHandler takes a CDO that must implement the CDO Interface as stated as an argument (line 8). After the document is parsed in line 11 the CDO can be retrieved again from the CMLHandler and used to access the data. Note that frames is the native object in JMol for the storage of all chemical data!
The internal structure of the handlers is the following. The Sax-events are passed to the root CMLHandler. The CMLHandler determines which convention should be used for this element and child element by checking the convention attribute. If there is a change in convention it dynamically loads a new convention handler, passes over all parsed data uptil then. In any case the next step will be passing on the event to the convention handler which might be new form then on. So the root handler does not actually process the CML file, it merely manages the processing. When a startElement event is raised it detects a change of convention and if necessary changes the convention handler.
Thus, this system always uses a convention handler that is specific for the convention given in the CML file. If no such convention handler is available it uses the default CML convention handler as pointed out by the DTD. At this moment, the convention handlers are hard coded, i.e. the source code of the CMLHandler needs to be changed when a new convention is to be supported.
However, it is easy to have the import filter use a plugin mechanism and dynamically loads convention handler as needed. This is not the case for JMol and JChemPaint at this moment, but this will be implemented in the near future. This plugin mechanisme makes it possible to add convention handlers without really upgrading JMol, JChemPaint or any other software that uses the CML import filter.